1. Get example data
library(tidyverse)
library(rstatix)
example <-
tibble::tibble(
para = rnorm(n = 30, mean = 300, sd = 20) %>% round(digits = 2),
dose = rep(rep(c("dose_1", "dose_2", "dose_3", "dose_4", "dose_5"), each = 2), 3),
date = rep(c("date_1", "date_2", "date_3"), each = 10)) %>%
group_by(dose, date) %>%
mutate(mean = mean(para)) %>%
mutate(sd = sd(para)) %>%
mutate(cv = sd/mean)
example
## # A tibble: 30 × 6
## # Groups: dose, date [15]
## para dose date mean sd cv
## <dbl> <chr> <chr> <dbl> <dbl> <dbl>
## 1 286. dose_1 date_1 289. 4.84 0.0167
## 2 293. dose_1 date_1 289. 4.84 0.0167
## 3 290. dose_2 date_1 299. 13.6 0.0455
## 4 309. dose_2 date_1 299. 13.6 0.0455
## 5 277. dose_3 date_1 303. 37.0 0.122
## 6 329. dose_3 date_1 303. 37.0 0.122
## 7 300. dose_4 date_1 307. 9.31 0.0303
## 8 313. dose_4 date_1 307. 9.31 0.0303
## 9 306. dose_5 date_1 286. 28.7 0.100
## 10 265. dose_5 date_1 286. 28.7 0.100
## # ℹ 20 more rows
2. Using Rstatix to calculate pvalue
# Using Rstatix to calculate pvalue
stat.test <- example %>%
rename("value" = colnames(.)[1]) %>%
filter(sd != 0) %>%
group_by(dose) %>%
mutate(x = n()) %>%
filter(x >= 4) %>%
t_test(value ~ date) %>%
filter(group1 == "date_1")
stat.test
## # A tibble: 10 × 11
## dose .y. group1 group2 n1 n2 statistic df p p.adj
## <chr> <chr> <chr> <chr> <int> <int> <dbl> <dbl> <dbl> <dbl>
## 1 dose_1 value date_1 date_2 2 2 -4.45 1.92 0.051 0.121
## 2 dose_1 value date_1 date_3 2 2 -7.81 1.40 0.04 0.121
## 3 dose_2 value date_1 date_2 2 2 1.03 1.77 0.424 1
## 4 dose_2 value date_1 date_3 2 2 -0.00833 1.84 0.994 1
## 5 dose_3 value date_1 date_2 2 2 0.245 1.86 0.831 1
## 6 dose_3 value date_1 date_3 2 2 0.718 1.16 0.59 1
## 7 dose_4 value date_1 date_2 2 2 1.95 1.55 0.226 0.678
## 8 dose_4 value date_1 date_3 2 2 0.107 1.09 0.931 1
## 9 dose_5 value date_1 date_2 2 2 -1.30 1.19 0.39 0.78
## 10 dose_5 value date_1 date_3 2 2 -0.462 1.14 0.717 0.78
## # ℹ 1 more variable: p.adj.signif <chr>
3. adjust lable position based on dodge width
stat.test <- stat.test %>%
add_xy_position(x = "dose", dodge = 0.75)
stat.test
## # A tibble: 10 × 16
## dose .y. group1 group2 n1 n2 statistic df p p.adj
## <chr> <chr> <chr> <chr> <int> <int> <dbl> <dbl> <dbl> <dbl>
## 1 dose_1 value date_1 date_2 2 2 -4.45 1.92 0.051 0.121
## 2 dose_1 value date_1 date_3 2 2 -7.81 1.40 0.04 0.121
## 3 dose_2 value date_1 date_2 2 2 1.03 1.77 0.424 1
## 4 dose_2 value date_1 date_3 2 2 -0.00833 1.84 0.994 1
## 5 dose_3 value date_1 date_2 2 2 0.245 1.86 0.831 1
## 6 dose_3 value date_1 date_3 2 2 0.718 1.16 0.59 1
## 7 dose_4 value date_1 date_2 2 2 1.95 1.55 0.226 0.678
## 8 dose_4 value date_1 date_3 2 2 0.107 1.09 0.931 1
## 9 dose_5 value date_1 date_2 2 2 -1.30 1.19 0.39 0.78
## 10 dose_5 value date_1 date_3 2 2 -0.462 1.14 0.717 0.78
## # ℹ 6 more variables: p.adj.signif <chr>, y.position <dbl>,
## # groups <named list>, x <dbl>, xmin <dbl>, xmax <dbl>
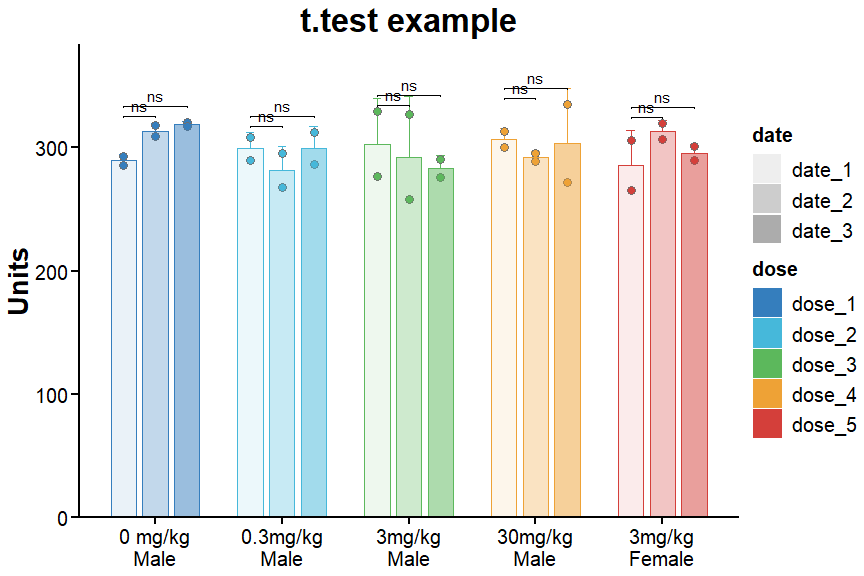
5. Get the final plot
ggplot(example, aes(x = dose, y = para, color = dose, fill = dose)) +
geom_errorbar(aes(ymin = mean, ymax=mean + sd, alph = date), width=.2,
position=position_dodge(.75), linewidth = 0.5) +
geom_point(aes(fil = date), shape = 21, size = 3, color = "gray44",
position = position_jitterdodge(dodge.width = 0.75, jitter.width = 0)) +
geom_bar(data = example %>% select(-1) %>% distinct(),
aes(y = mean, fill = dose, color = dose, alpha = date),
stat = "identity", width = 0.6, position = position_dodge(width = 0.75)) +
scale_color_manual(values = ggsci::pal_locuszoom()(5) %>% rev()) +
scale_fill_manual(values = ggsci::pal_locuszoom()(5) %>% rev()) +
scale_alpha_manual(values = c(seq(1, 9, 2))*0.1) +
scale_y_continuous(expand = expansion(mult = c(0, 0.1)), name = "Units") +
# scale_y_continuous(name = plot_data %>% colnames() %>% nth(n = 1) %>% str_remove(pattern = "\\|.*\\|"),
# expand = c(0, 0), limits = c(0, max(plot_data %>% pull(1))*1.1)) +
labs(title = "t.test example") + xlab(NULL) +
cowplot::theme_cowplot(font_size = 10, line_size = 0.8) +
theme(axis.title = element_text(size = 20, face = "bold"),
axis.text = element_text(size = 15, color = "black"),
axis.ticks.length = unit(.2, "cm"),
plot.title = element_text(size = 25, hjust = 0.5, face = "bold"),
legend.key.size = unit(0.8, "cm"),
axis.line = element_line(colour = "black", size = 1),
# panel.border = element_rect(size = 1, colour = "black"),
axis.ticks = element_line(colour = "black", size = 1),
legend.text = element_text(size = 15),
plot.caption = element_text(size = 12, hjust = 1.2, face = "italic"),
legend.title = element_text(size = 15, face = "bold"),
strip.text = element_text(face = "bold", size = 15)) +
ggpubr::stat_pvalue_manual(stat.test, label = "p.adj.signif", tip.length = 0.01, bracket.nudge.y = 0) +
scale_x_discrete(labels = c("dose_1" = "0 mg/kg\nMale",
"dose_2" = "0.3mg/kg\nMale",
"dose_3" = "3mg/kg\nMale",
"dose_4" = "30mg/kg\nMale",
"dose_5" = "3mg/kg\nFemale"))