此post旨在提取heatmap作图中的cluster行名或者列名
其实,对heatmap热图提取cluster就是对hclust的结果提取cluster,tree_col和tree_row属性
首先,我们要构建一个matrix或者data.frame进行pheatmap作图
# package library
library(pheatmap)
library(tidyverse)
# Create random data
set.seed(50)
data <- replicate(20, rnorm(50, mean = 100, sd = 100))
data %>% class()
rownames(data) <- paste("Gene", c(1:nrow(data)))
colnames(data) <- paste("Sample", c(1:ncol(data)))
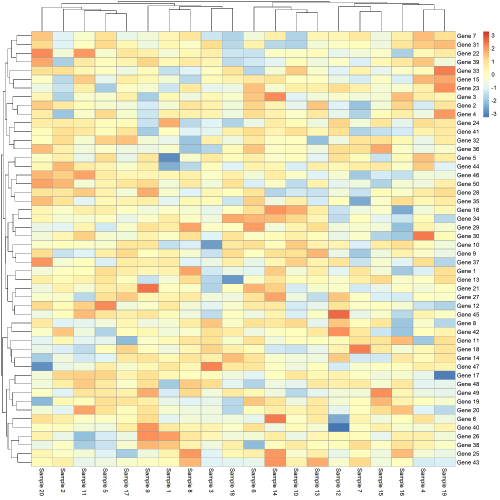
# get heatmap plot
(out <- pheatmap(data, scale="row"))
dev.copy(png, "heatmap_raw.png", width=4000, height=4000, res = 330)
dev.off()
随后,我们提取热图中的行名和列名
# Re-order original data (genes) to match ordering in heatmap (top-to-bottom)
data[out$tree_row[["order"]],] %>% rownames()
# Re-order original data (samples) to match ordering in heatmap (left-to-right)
data[out$tree_row[["order"]],] %>% colnames()
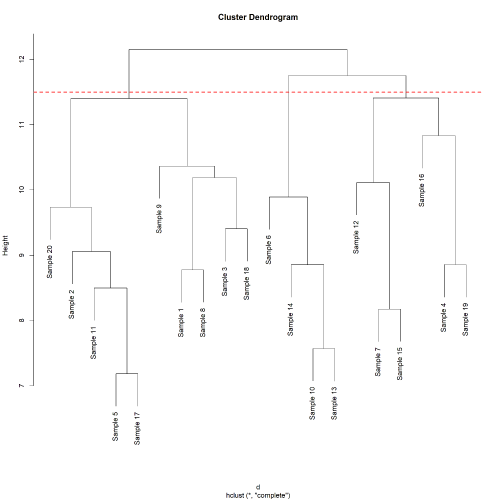
# get hclust plot
plot(out$tree_col)
abline(h = 11.5, col="red", lty=2, lwd=2)
dev.copy(png, "heatmap_hclust.png", width=4000, height=4000, res = 330)
dev.off()
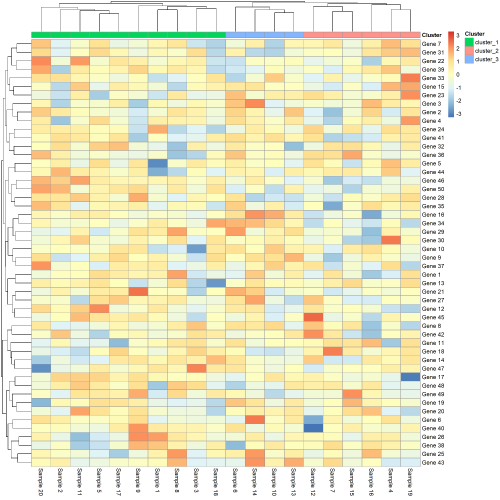
假如我们需要可视化的话,可以加一个annotation
# get 3 cluster for sample name
(tree <- cutree(out$tree_col, k=3))
tree <- ifelse(tree == 1, "cluster_1",
ifelse(tree == 2, "cluster_2",
"cluster_3")) %>% as.data.frame() %>% setNames("Cluster")
# get annotation plot
pheatmap(data, scale = "row", annotation_col = tree)
dev.copy(png, "heatmap_3cluster.png", width=4000, height=4000, res = 330)
dev.off()